Submitted by Michael Dondrup on
Frequently Asked Questions
Q: I want to check if I added one of my RNAi experiments, and don't want to look through all that has been added. How do I search by using title and/or target values? For me both options would be appreciated.
A: It is easiest to search titles of all documents on this site using the "Search this site", simply enter parts of the title and press search. More complex search forms are available under "RNAi" - "browse by target". You can also create a bookmark in your browser if you work with a single experiment often.
You can create a shortcut by editing and adding your name to the experiment under General - Experiment contacts. Then, the experiment will show up on your personal account page each time you log in or when you go to "My account".
Q: And what is the gene symbol? What do you mean? What parameter should I use?
A: The gene symbol is deprecated and should no longer be used in favor of the stable id; it was used during a time when we didn't have a genome annotation, to provide a hint to what the intended target should be. You can still search for it for backwards compatibility, because there are several experiments which do not have a stable ID yet.
Q: I work with a lot of genes. One example is a gene of about 6500bp and it does not have an Ensembl stable ID. It needs to have one. The gene has been verified. Could you please help me with that?
A: We cannot provide you with an Ensembl stable ID immediately because these are assigned by Ensembl. Please do not try to make a 'fake Ensembl' id by naming your gene. Instead follow our gene naming conventions. But, you can obtain a preliminary ID by submitting your sequence. If you are an annotator, you can upload your sequence yourself and use it immediately in your RNAi experiment. Please follow the instructions under
If you are not an annotator, you need to contact an annotator or the administrator, after filing a
because you cannot upload a new feature yourself if you are not an annotator. The gene will possibly get assigned a new Ensembl stable id in the next release if the evidence you provide is convincing.
Q: Another one of my genes are probably not correct. I have a stable-ID. I tried BLAST in a lot of variations with the amino-acid and DNA sequence from my stable-ID gene. Why can I only BLAST against genomic sequence and not predicted genes? Or how do I do it? I want to check that the ID is correct, and if my new sequence will give any hits.
A: You can run Blast against the predicted genes. Select the Blast database called "L. salmonis Ensembl predicted proteins" using program: Blastp. If you do not find the gene this way, it has been probably missed by the automatic annotation. In this case you can use these options:
- Input: DNA sequence, Program: Tblastx, Database: 1. "L. salmonis scaffolds (masked)", and then 2."Everything L.salmonis" if you don't find it in 1.
- Input: AA sequence, Program: Tblastn, Database: 1. "L. salmonis scaffolds (masked)", and then 2."Everything L.salmonis" if you don't find it in 1.
If you find the sequence, you can use the link "Show in GBrowse" to see if the sequence hit overlaps with known genes.
Q: I have obviously made a mistake and now everything is locked. Wrote something in the wrong place, and now I’m not able to get back to zero. This turns up every time I try to open “Browse RNAi by target” Other stuff seems to work. Have tried to log out and in. Did not work
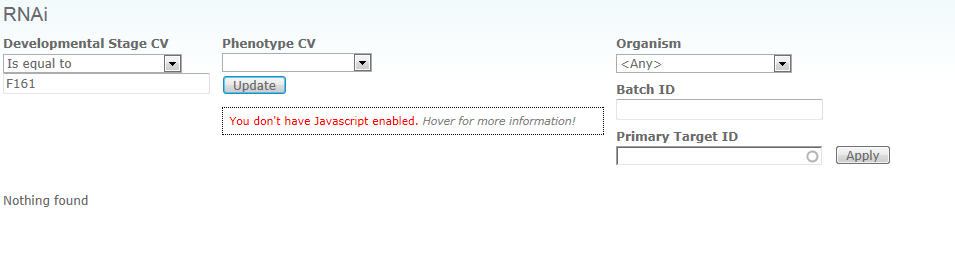
A: you should be able to simply clear the field Developmental Stage CV, and the form should forget the value you entered initially. You may also ignore the "You don't have Javascript enabled" warning, if you know you have Javascript enabled. It will sometimes show up when you hit the reload button on this page.
GBrowse questions
Q: I am trying to open an protein sequence (EMLSAP...) in GBrowse, but it doesn't show me any features and the 6-frame translation track shows an error ("Cannot translate amino-acid sequence").
A: You can find all protein sequences in GBrowse but you can't do much useful with them for now. The translation error is to be expected because AA sequence is already translated. We might provide protein domain annotation on the AA-sequence in the future.
Blast questions
Q: I have run BlastN on a nucleotide sequence against the genome and got a hit which is shown to overlap a gene. I exported the gene sequence from LiceBase and Ensembl, but I cannot find my query sequence in either. Which is wrong, Blast or the gene sequence?
A: They are both correct. However, overlaps in our BlastN report are given without taking the strand information of the genes into account. Possibly the gene is on the opposite strand of the match (e.g. match on + strand, gene on -). LiceBase and Ensembl report the gene sequence in the correct way with respect to the gene strand. In this case you need to reverse-complement either the gene sequence or the query and you will find the sequence.
| Attachment | Size |
|---|---|
| 15.86 KB |
