Submitted by Administrator on
The comprehensive guide to finding your Gene of Interest
How you can search your genes depends on what you have as input. We can differentiate two main cases:
- Searching genes by annotation (gene name, ID, description, EC-number, GO-term)
- Searching genes by their sequence (nucleotide or amino acid sequence)
There are likely always multiple ways to discover genes, and often there is not a single best way (yeah, genomics is complex). However, if you looking for the easies way to get started, try
In most cases it is not necessary to blast an external sequence, because it has already been done before.
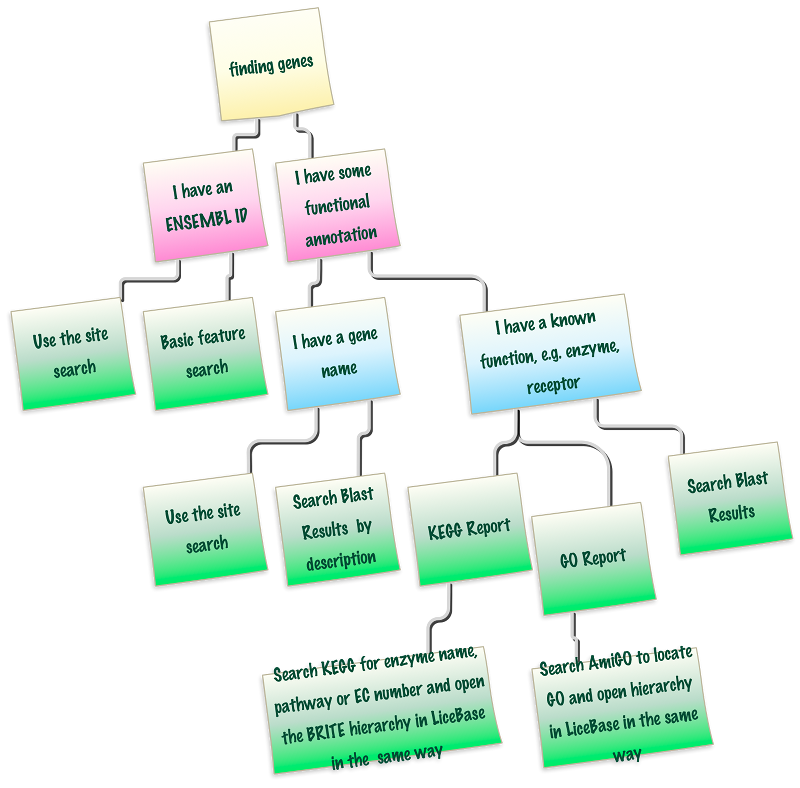
Finding genes from functional annotation
Out of the two cases, finding genes by annotation is often the most straight-forward approach if the gene is already annotated. You might, for example look for an EMLSAG ID directly, or you could search directly for "chitin synthase". See the overview graph below, to help with your decision. In many cases, searching the pre-computed blast results is very convenient to get a quick overview. In any case, all other evidence should be checked, for the gene found, to confirm it is really the gene you are looking for.
Hint: the graph is linked to the correct pages, green notes are clickable directly.
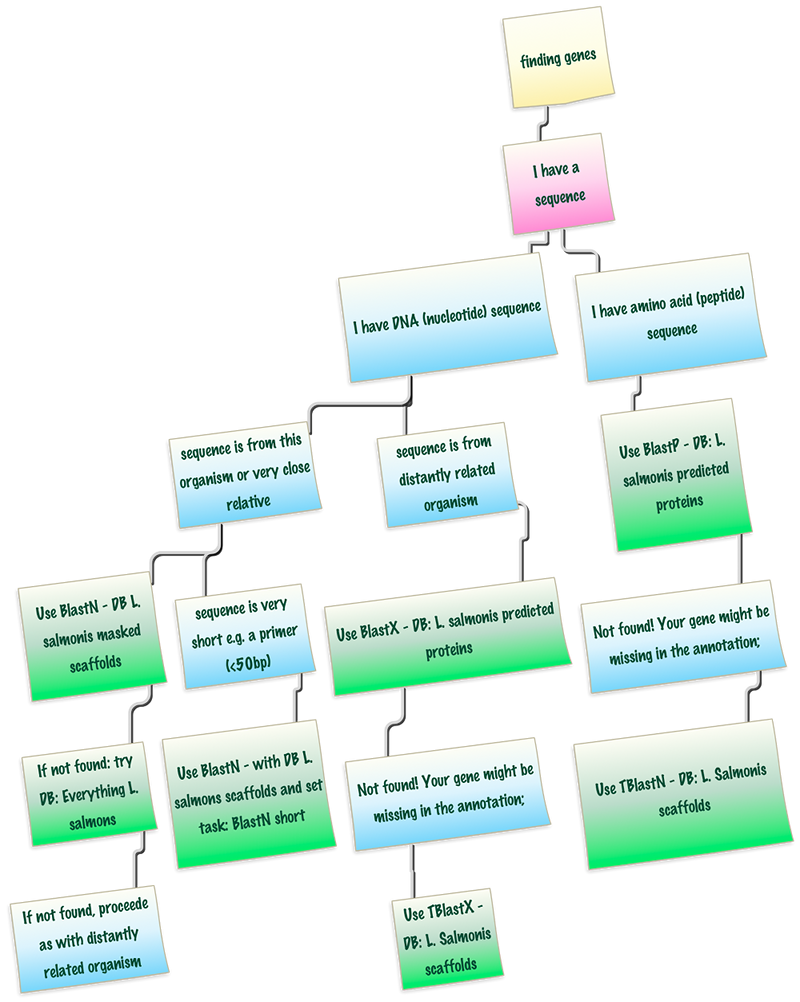
Searching genes by sequence
Sometimes, there might be only sequence information available to search for a gene. We differentiate two cases:
- DNA-sequence, e.g. the result of PCR amplification and sequencing of a gene in this organism
- AA-sequence, e.g. the protein sequence of the gene of interest from another organism.
For DNA sequences we also need to know if they are from the organism in question or a closely related one, or if they are from a more distant organism. In this case they need to be treated differently. All search options involve different Blast tools (BlastN, BlastP, BlastX, etc.). Please check the blast options carefully and especially the blast database.
Hint: the graph is linked to the correct pages, green notes are clickable and will open a browser tab with the correct Blast options pre-selected! Simply paste the search sequence there.
What if I cannot find the gene that way?
If you have tried every method above and still haven't found the gene or the class of gene you are looking, don't give up. There are several more sensitive methods such as PSI-Blast and HMMER tools. They are just not integrated into LiceBase (yet) due to computational requirements. Just contact us. Certainly, there will be a certain amount of genes that are really missing in the salmon louse, but if a gene or gene family is present, we will also discover it.
