Submitted by Administrator on
It's just a web-site
LiceBase is just a website. As such it consists of web-pages and hyperlinks between them. There are also web-forms for data entry and searches. As with most web-sites how to use them should be more or less self-explanatory. Also, most explanations will be embedded into the web-pages, e.g. into web-forms like shown below:
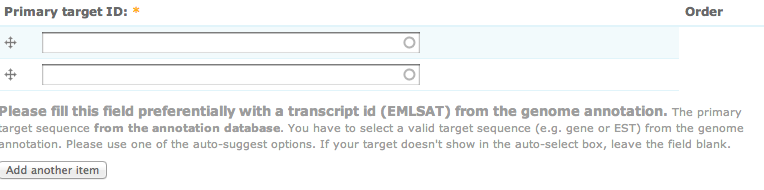
Web-sites also change over time as more content is added. That is the same here, so a little bit of exploration is required.
Bookmarks and links work normal, so that you can link to almost any page in LiceBase.
- you can link to from a document or web-page or bookmark a gene like so: https://licebase.org/EMLSAG00000000001
- for the same gene in GBrowse you can bookmark a link like this: https://licebase.org/fgb2/gbrowse/lsalmonis/?q=EMLSAG00000000001
It's an account based web-application
LiceBase is not a static web-site, but a complex bundle of web-applications (just like Facebook or Google+). You can interact with the site, make new content and annotate genes and experiments if you have an account. You can also write a Blog or ask questions as Forum entries. You can also write your own documentation.
Some important points to note:
- You need to login to get full access to all functions. The site can look different if you are not logged in.
- Most parts of the site is not indexed by Google of other search engines. You can use the site-search instead:
- You can contribute actively by using "Web 2.0 features" like writing Blog posts, Forum entries, etc.
We are using established standard software, namely Drupal and GMOD tools to build LiceBase.
- Drupal is a content management system that is highly configurable and allows to develop very powerful web-tools. It has a huge amount of features and enables
- Tripal is a collection of extension modules for Drupal to create genomics oriented web-sites (generates Gene and Organism pages)
- GBrowse is a web-based genome browser. GBrowse comes with its own user documentation.
- NCBI Blast+ commandline tools are used for sequence search (not the web interface)
- The web interface to Blast is based on the GMOD component Blast Graphic Viewer
