Submitted by Administrator on
Repetitive sequences are abundant in most eukaryote genomes. They pose difficulties for an exact reconstruction of the genomic sequence and also for primer design. They can also be useful as genetic markers, as micro- or mini-satelites.
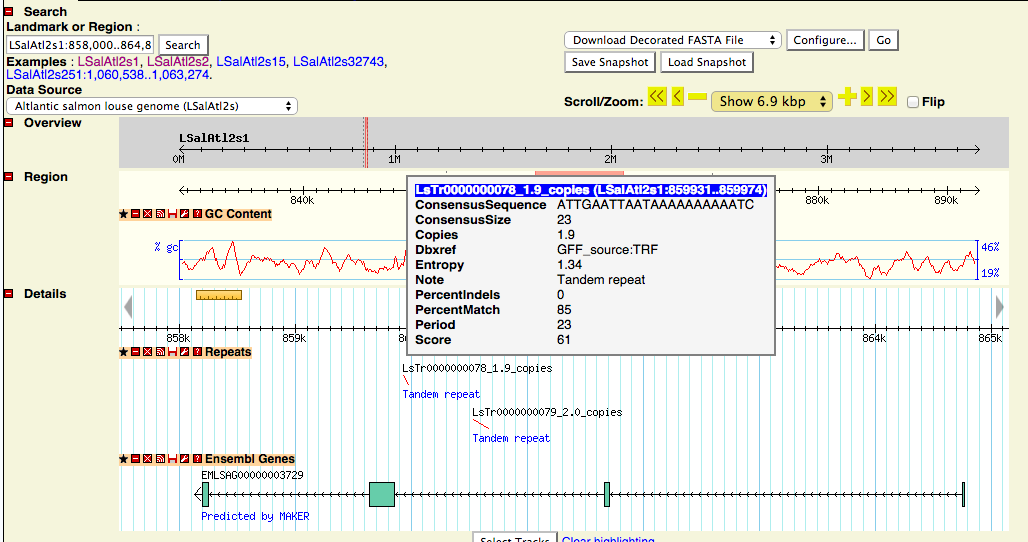
Often it is impossible to determine the exact size of such inserts, a short tandem repeat might be visible with only 2 copies, for example, but in fact stretch a region of several kilo bases. This can be a problem for PCR resulting in failure to produce a product or product of unexpected size, because the DNA sequence containing a repeat can be much larger than visible at first glance.
Therefore, I have created a new track in the GenomeBrowser, called "Repeats". Currently, this track contains only tandem repeats from Tandem Repeats Finder. Tandem repeats (TR) are mainly present in introns and intergenic regions. They are very common, only very few contigs, including the mitochondrial genome do not contain any TR. TR can conceal a large insert of repeat sequence, even though their average periodicity in the genomic sequence is approximately only 2. There will be added a track with more different types of repeats soon.
In summary, we recommend that a PCR-primer pair should not span a tandem repeat region, or the PCR might fail.
A Gene with intronic tandem repeats. On mouse-over, more information about the repeat is revealed.
| Attachment | Size |
|---|---|
| 89.11 KB |
