Tigriopus kingsejongensis
|
Overview
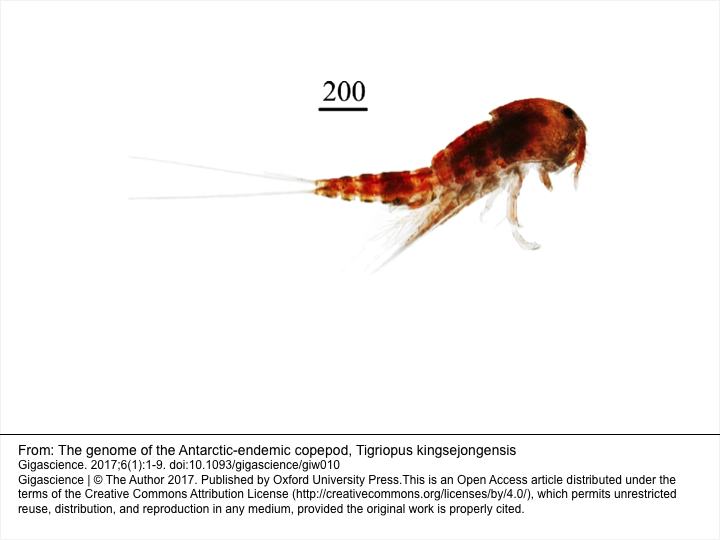 Tigriopus kingsejongensis is a free-living intertidal copepod endemic to the Antarctic. The T. kingsejongensis genome has been recently published and provides an interesting example of cold adaptation [1]. The genome annotation was done using the Maker pipeline. The T. kingsejongensis genome and annotation have been imported into LiceBase from public data provided with the original publication and orthologs between the species have been computed by BlastP based on the automatic annotations. 1: Kang S, Ahn DH, Lee JH, Lee SG, Shin SC, Lee J, Min GS, Lee H, Kim HW, Kim S, Park H. The genome of the Antarctic-endemic copepod, Tigriopus kingsejongensis. Gigascience. 2017 Jan 1;6(1):1-9. doi: 10.1093/gigascience/giw010. PubMed PMID: 28369352; PubMed Central PMCID: PMC5467011. Feature Summary
The following features are currently present for this organism
Feature Browser
The following browser provides a quick view for new visitors. Use the searching mechanism to find specific features.
PagesCross References
External references for this organism
Downloads
|
